Protein Refolding
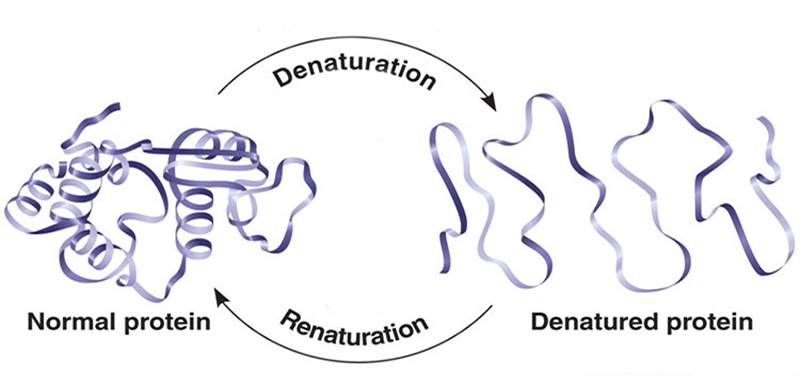
One of the main stumbling blocks encountered when attempting to express heterologous proteins in Escherichia coli is the accumulation of inactive and improperly folded proteins in the form of aggregates, called inclusion bodies (IB). Even though several approaches have been used to mitigate protein misfolding during overexpression of proteins in E. coli, over 70% of recombinant proteins expressed in E. coli are contained in inclusion bodies. Developing efficient protein native structure recovery procedures based on IB refolding is therefore an important challenge. Different techniques have been designed to recover correctly folded proteins from inclusion bodies including dilution, dialysis and chromatography, etc.
Techniques of protein refolding
| Refolding techniques | Descriptions | Features | |
|---|---|---|---|
| Dilution | Fast dilute inclusion body protein solution with refolding buffer, achieving the goal of lowering the concentration of denaturant, then making a refolded protein. |
|
|
| Dialysis | Relatively slow removal of denaturant by buffer exchange through a membrane of defined molecular weight cut-off. |
|
|
| Refolding by chromatography | Gel filtration chromatography | Molecular sieve effect can separate the proteins and small molecules denaturant, achieving solution exchange and protein refolding. |
|
| Adsorption chromatography (SEC、HIC、IEC、IMAC) | The adsorption of denatured protein on a medium can effectively inhibit non-specific interactions between protein molecules, so as to improve protein refolding rate. | ||
| Chromatography refolding mediated by immobilized chaperone and/ or foldase | Immobilized chaperone or foldase on medium makes the chromatographic column a folding reactor. | ||
| Refolding by reverse micelle | Protein molecules can be wrapped in reverse micelle, reducing the aggregation in the process of protein folding, and through gradually reducing the concentration of the denaturant and joining the REDOX, denatured proteins refold. |
|
|
| Refolding by aqueous two-phase extraction | Aqueous two-phase system makes the dissolution of inclusion bodies and protein refolding completed in one step. |
|
|
| Refolding by high hydrostatic pressure | High pressure can solubilize aggregates while favoring the native protein conformation. |
|
|
Refolding additives
Protein refolding from denatured proteins is influenced by several factors, including solubility of protein, removal of denaturant, and assistance of refolding additives. The addition of chemical additives has been frequently used to prevent protein aggregation. Chemical additives can have effects in the following aspects: stabilizing the natural structure of correct folded proteins, changing the stability of misfolded proteins, increasing the solubility of folding intermediates and un-folded molecules. Commonly used chemical additives are shown below:
Denaturants: urea and GdnHCl
Amino acids: Gly, Ala, Pro, and Arg
Polymers: PEG and cyclodextrin
Polyols: glycerol
Alcohol: short-chain alcohols
Salts: NaCl, KCl, MgCl2 and (NH4)2SO4
Sugars: glucose, sucrose and trehalose
Detergents: Triton X-100, CHAPS, SDS and CTAB Detergents with cycloamylose or cyclodextran
Artificial chaperones: thioredoxin, disulfide bond isomerase
Reducing/oxidizing reagents: DTT, cysteine/cystine and glutathione
Non-detergent zwitterionic agents: Non-detergent sulfobetaines (NDSB)
Though different techniques have developed for the generation of correctly folded protein from solubilized inclusion bodies, there is no universal refolding recipe for any protein. Proteins folding in solution are affected by a number of physiochemical parameters including ionic strength, pH, temperature, oxidation state and protein concentration as well as the presence of hydrophobic, polar, chaotropic agents and other proteins. For any given protein, the best refolding conditions still have to be determined empirically. BiologicsCorp has rich experiences to solve problems of protein refolding in E.coli. After evaluation of target proteins' biochemical and the structural properties, BiologicsCorp can offer flexible mature solutions to solve problems of protein refolding.
Detection of protein refolding efficiency
According to specific protein properties, the efficiency of protein refolding can be tested from the aspects of biochemical, immune, physical.
- By functional assay: it is best to perform a functional assay to determine if any active protein is present.
- By immunoassay: by performing an immunoblot or ELISA, antibody test of structure determinant can reflect the real state of protein folding.
- By SDS-PAGE: successful refolding is evidenced by the presence of the target protein in the liquid fraction.
- By size exclusion chromatography: misfolded or aggregated protein will have a different retention time than correctly folded protein.
- By viscosity and turbidity measurement: protein solubility increases after renaturation, protein in denaturation condition due to hydrophobic residues generally has poor solubility in water, mostly forming visible precipitate out.
FAQs about protein refolding
1.How best can I refold a recombinant protein from inclusion bodies and retain its bioactivity?
The methods normally used for solubilization of inclusion body could lead to non-native conformation of the expressed protein. This problem could be resolved by proper refolding procedures of target protein at low denaturant concentrations. Higher concentration of the unfolded protein often leads to decreased refolding yields, regardless of refolding method. So, it is desirable to keep the concentration of the initial un-folded protein to a minimum level if higher and correct refolding proteins are expected.
2.How to choose protein concentration when renaturation?
Generally, initial protein concentration is 0.1-1 mg/ml, high concentration is easy to form a dimer precipitation, low concentration is not economic , and many kinds of proteins in low concentration is not stable, easy to denature.
3.What is the reason of protein precipitation during dialysis? How to deal with this problem?
Protein precipitation during dialysis against phosphate or Tris buffers can be caused by: too low salt concentration, too high protein concentration, sudden pH changes and pH of dialysis buffer close to PI of a protein. Therefore, it is advisable to use at least 1000 fold excess of dialysis buffer, perform procedure at 4℃(at least 3 hours, followed by a buffer change), use at least 350 mM salt in the dialysis buffer, 50 % glycerol is optional. Besides, precipitated proteins are very difficult to refold. If possible, purify your protein again and apply other conditions for dialysis.
Case study
Refolding technology in dimeric protein (disulfide bond):
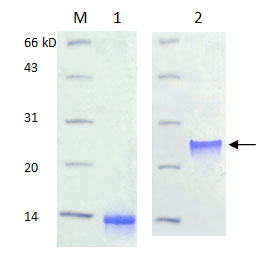
Lane M: Protein maker
Lane 1 : Before refolding-Protein B201 expressed as inclusion body and no activity in monomer.
Lane 2 : After refolding-High biological activity in dimer connected by disulfidebond.
Related information
2.How can we reduce or avoid protein misfolding during heterologous protein over expression?
Optimization of soluble protein expression is often the strategy of choice when trying to obtain bioactive protein. Several approaches have been used to mitigate protein misfolding during over expression of proteins in E. coli. These include: 1) fusion of the target protein with a more soluble partner, typically a bacterial protein; 2) co-expression of folding catalysts and chaperones; 3) expression under culture conditions which reduce the translation rates or affect the intracellular environment; and 4) modification of the protein sequence. Each approach has advantages and disadvantages which must be weighed in light of the intended end-use of the target protein. Further, not all proteins respond favorably to any given approach. Again, which approach is best suited to a given protein must be determined empirically and success in producing and recovering soluble active protein is not guaranteed. The formation of inclusion bodies is still common and often unavoidable.
References
De Bernardez-Clark E: Refolding of recombinant proteins. Current Opinion in Biotechnology, 1998, 9: 157–163.
Tsumoto K, Ejima D, Kumagai I, Arakawa T: Practical considerations in refolding proteins from inclusion bodies. Protein Expression and Purification, 2003, 28: 1-8.
Luis Felipe Vallejo, Ursula Rinas: Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microbial Cell Factories, 2004, 3(1):85-100.
Gu Z, Weidenhaupt M, Ivanova N, Pavlov M, Xu B, Su Z-G, Janson JC: Chromatographic methods for the isolation and refolding of proteins from Escherichia coli inclusion bodies. Protein Expression and Purification, 2002, 25: 174-179.
Werner MH, Clore GM, Gronenborn AM, Kondoh A, Fisher RJ: Refolding proteins by gel filtration chromatography. FEBS Letters, 1994, 345: 125-130.
Contact Us


