Protein Expression and Purification

Overview of protein expression and purification
What do you know about your protein
How to improve the expression level of active and soluble protein
Strategies for native protein and recombinant protein purification
Methods for protein seperation and protein purification
Custom Protein Service & Contact Us


Which protein expression system suits your needs?
 What systems are currently used in the laboratory or by others in the field? If the protein coding sequence of interest is well characterized, and the protein or its close relatives have been expressed successfully by others in the field, it is wise to try the same gene expression system. Go with what has worked in the past. If nothing else, results obtained using the familiar system will serve as a starting point. As an example, most of the recombinant protein expression of mammalian src homology SH2 protein interaction domains has been done using the pGEX vector series, and similar examples of preferred systems are found in other fields of research. If little is known about the protein to be expressed, it is best to take stock of what information there is before entering the lab. Before beginning any experimentation, it is wise to answer the following question:
What systems are currently used in the laboratory or by others in the field? If the protein coding sequence of interest is well characterized, and the protein or its close relatives have been expressed successfully by others in the field, it is wise to try the same gene expression system. Go with what has worked in the past. If nothing else, results obtained using the familiar system will serve as a starting point. As an example, most of the recombinant protein expression of mammalian src homology SH2 protein interaction domains has been done using the pGEX vector series, and similar examples of preferred systems are found in other fields of research. If little is known about the protein to be expressed, it is best to take stock of what information there is before entering the lab. Before beginning any experimentation, it is wise to answer the following question:
WHAT DO YOU KNOW ABOUT YOUR GENE?
Source
In general, simple globular proteins from prokaryotic and eukaryotic sources are good candidates for gene expression in E. coli. Monomeric proteins with few cysteines or prosthetic groups (e.g., heme and metals) and of average size (<60kDa) will likely give good production. Secreted eukaryotic proteins and membranebound proteins, especially those with several transmembrane domains, are likely to be problematic in E. coli. Solubility of recombinant proteins in E. coli can also be estimated by a mathematical analysis of the amino acid sequences (Wilkinson and Harrison, 1991).
Codon Usage
Codon usage may also affect the level of protein expression. If the gene of interest contains codons not commonly used in E. coli, low expression may result due to the depletion of tRNAs for the rarer codons. When one or more rare codons is encountered, translational pausing may result, slowing the rate of protein synthesis and exposing the mRNA to degradation. This potential problem is of particular concern when the sequence encodes a protein >60kDa, when rare codons are found at high frequency, or when multiple rare codons are found over a short distance of the coding sequence. For example, rare codons for arginine found in tandem can create a recognition sequence for ribosome binding (e.g., _AGGAGG) that closely approximates a Shine- Dalgarno sequence UAAGGAGG.This may bind ribosomes nonproductively and block translation from the bona fide ribosome binding site (RBS) at the initiator codon further upstream. Nonetheless, the appearance of a rare codon does not necessarily lead to poor expression. It is best to try the native gene expression, and then make changes if these seem warranted later. Strategies include mutating the gene of interest to use optimal codons for the host organism, and co-transforming the host with rare tRNA genes. In one example, introduction into the E. coli host of a rare arginine (AGG) tRNA resulted in a several-fold increase in the expression of a protein that uses the AGG codon (Hua et al., 1994). In another case, substitution of the rare arginine codon AGG with the E. coli-preferred CGU improved expression (Robinson et al., 1984). Other work has shown that rare codons account for decreased expression of the gene of interest in E. coli (Zhang, Zubay, and Goldman, 1991; Sorensen, Kurland, and Pederson, 1989). Rare codons may have an even more dramatic.
Secondary Structure
 Secondary structures that occur near the start codon may block translation initiation (Gold et al., 1981; Buell et al., 1985), or serve as translation pause sites resulting in premature termination and truncated protein. These can be found using DNA or RNA analysis software. Structures with clear stem structures greater than eight bases long may be disrupted by site-specific mutation or by making all or a portion of the coding sequence synthetically. Depending on the size of the gene, and the importance of obtaining high-expression levels, it may be worth synthesizing the gene. This has been generally done by synthesizing overlapping oligonucleotides that when annealed can be extended using PCR and ligated to form the full-length coding sequence. There are several examples where this approach has been used to optimize codon usage for E. coli (Koshiba et al., 1999; Beck von Bodman et al., 1986). In addition, if one takes on the work and expense of synthesizing a gene, secondary structures in the predicted RNA that might stall translation can be removed, and sites for restriction endonucleases can be introduced. Coding sequences with high GC (>70%) content may reduce the level of expression of a protein in E. coli. Check the sequence using a DNA analysis program.
Secondary structures that occur near the start codon may block translation initiation (Gold et al., 1981; Buell et al., 1985), or serve as translation pause sites resulting in premature termination and truncated protein. These can be found using DNA or RNA analysis software. Structures with clear stem structures greater than eight bases long may be disrupted by site-specific mutation or by making all or a portion of the coding sequence synthetically. Depending on the size of the gene, and the importance of obtaining high-expression levels, it may be worth synthesizing the gene. This has been generally done by synthesizing overlapping oligonucleotides that when annealed can be extended using PCR and ligated to form the full-length coding sequence. There are several examples where this approach has been used to optimize codon usage for E. coli (Koshiba et al., 1999; Beck von Bodman et al., 1986). In addition, if one takes on the work and expense of synthesizing a gene, secondary structures in the predicted RNA that might stall translation can be removed, and sites for restriction endonucleases can be introduced. Coding sequences with high GC (>70%) content may reduce the level of expression of a protein in E. coli. Check the sequence using a DNA analysis program.
Size of a Gene or Protein
 As a rule, very large (>100kDa) and very small (<5kDa) proteins are more difficult to express in E. coli. Small polypeptides with little secondary structure tend to be rapidly degraded in E. coli. Degradation can be minimized by expressing such short oligopeptides as concatemers with proteolytic or chemical cleavage sites in between the monomeric units (Hostomsky, Smrt, and Paces, 1985). Short peptides are also successfully expressed as fusion proteins. Fusion with GST, MalB or other larger, well folded partners will tend to stabilize a short peptide, making expression possible and purification relatively simple. One publication has shown MBP to be superior to other large fusion proteins at stabilizing short polypeptides (Kapust and Waugh, 1999). At the other extreme, proteins that are above 60kDa are best made using smaller affinity tags, such as FLAG, his6, or on their own, without any fusion. While there is no clear upper limit, the larger the protein, the lower the yield is likely to be.
As a rule, very large (>100kDa) and very small (<5kDa) proteins are more difficult to express in E. coli. Small polypeptides with little secondary structure tend to be rapidly degraded in E. coli. Degradation can be minimized by expressing such short oligopeptides as concatemers with proteolytic or chemical cleavage sites in between the monomeric units (Hostomsky, Smrt, and Paces, 1985). Short peptides are also successfully expressed as fusion proteins. Fusion with GST, MalB or other larger, well folded partners will tend to stabilize a short peptide, making expression possible and purification relatively simple. One publication has shown MBP to be superior to other large fusion proteins at stabilizing short polypeptides (Kapust and Waugh, 1999). At the other extreme, proteins that are above 60kDa are best made using smaller affinity tags, such as FLAG, his6, or on their own, without any fusion. While there is no clear upper limit, the larger the protein, the lower the yield is likely to be.
WHAT DO YOU KNOW ABOUT YOUR PROTEIN?
Cysteines
 There are many things that E. coli does not do well, or at all. If the protein of interest is naturally multimeric, or requires posttranslational modifications for activity, E. coli as an gene expression host may be a poor choice. Disulfide bonds, formed between two cysteines in an expressed protein, are made inefficiently in the reducing environment of the E. coli cytoplasm (Bessette et al., 1999; Derman et al., 1993). If the protein is produced, and can be purified from E. coli, in vitro oxidation of the cysteines may be tried (Dodd et al., 1995). Alternatively, the gene of interest can be cloned in a vector that includes a signal sequence (e.g., OmpA, geneIII, and phoA) that will direct the recombinant protein to the relatively oxidizing environment of the periplasm of E. coli, where disulfide formation is more efficient. Strains of E. coli that are deficient in thioredoxin reductase (trxB) permit proper disulfide formation in the cytoplasm (Derman et al., 1993;Yasukawa et al., 1995). Subsequent work has produced strains that lack both trxB and glutathione oxidoreductase and give better rates of disulfide formation than those seen in native E. coli periplasm (Bessette et al., 1999).
There are many things that E. coli does not do well, or at all. If the protein of interest is naturally multimeric, or requires posttranslational modifications for activity, E. coli as an gene expression host may be a poor choice. Disulfide bonds, formed between two cysteines in an expressed protein, are made inefficiently in the reducing environment of the E. coli cytoplasm (Bessette et al., 1999; Derman et al., 1993). If the protein is produced, and can be purified from E. coli, in vitro oxidation of the cysteines may be tried (Dodd et al., 1995). Alternatively, the gene of interest can be cloned in a vector that includes a signal sequence (e.g., OmpA, geneIII, and phoA) that will direct the recombinant protein to the relatively oxidizing environment of the periplasm of E. coli, where disulfide formation is more efficient. Strains of E. coli that are deficient in thioredoxin reductase (trxB) permit proper disulfide formation in the cytoplasm (Derman et al., 1993;Yasukawa et al., 1995). Subsequent work has produced strains that lack both trxB and glutathione oxidoreductase and give better rates of disulfide formation than those seen in native E. coli periplasm (Bessette et al., 1999).
Membrane Bound
 If the protein to be expressed is naturally associated with membrane and/or has at least one transmembrane domain, addition of a secretion signal to the amino terminus may help to maximize expression of functional protein. Signal sequences, about 20 residues long are derived from proteins that naturally are secreted into the periplasmic space, such as pelB, OmpA, OmpT, MalE, alkaline phosphatase (phoA), or geneIII of filamentous phage (Izard and Kendall, 1994). Protein with an amino terminal signal will be directed to the inner membrane of E. coli, and the carboxy terminal portion of the protein will be translocated into the periplasmic space. Depending on the hydrophobicity of the protein of interest, it may not translocate entirely into the periplasm but remain associated with the inner membrane. Secretion may help stabilize proteins from proteolytic attack (Pines and Inouye, 1999), or at least can reduce aggregation of hydrophobic proteins in the cytoplasm, and minimize inclusion body formation. Because of the reducing environment of the periplasmic space, proteins that contain one or more disulfide bonds are best secreted. The presence of an N-terminal signal sequence appears to be necessary but not sufficient to direct a target protein to the periplasm. Translocation across the outer membrane and into the growth medium is inefficient. In most cases target proteins found in the growth medium are the result of damage to the cell envelope and do not represent true secretion (Stader and Silhavy, 1990). Translocation across the inner cell membrane of E. coli is incompletely understood (reviewed by Wickner, Driessen, and Hartl, 1991), and the efficiency of export will depend on the individual target protein. Currently the export cannot be predicted based on protein sequence, although some generalizations have been made about the sequence immediately following the signal peptide (Boyd and Beckwith, 1990; Yamane and Mizushima, 1988). Therefore it is possible to find target proteins in the cytoplasm (with uncleaved signal sequence) or in the periplasm in partially processed form, in place of or in addition to the expected periplasmic processed species. In some cases the proportion of protein that is exported can be increased by lowering the temperature 15 to 30°C during induction.
If the protein to be expressed is naturally associated with membrane and/or has at least one transmembrane domain, addition of a secretion signal to the amino terminus may help to maximize expression of functional protein. Signal sequences, about 20 residues long are derived from proteins that naturally are secreted into the periplasmic space, such as pelB, OmpA, OmpT, MalE, alkaline phosphatase (phoA), or geneIII of filamentous phage (Izard and Kendall, 1994). Protein with an amino terminal signal will be directed to the inner membrane of E. coli, and the carboxy terminal portion of the protein will be translocated into the periplasmic space. Depending on the hydrophobicity of the protein of interest, it may not translocate entirely into the periplasm but remain associated with the inner membrane. Secretion may help stabilize proteins from proteolytic attack (Pines and Inouye, 1999), or at least can reduce aggregation of hydrophobic proteins in the cytoplasm, and minimize inclusion body formation. Because of the reducing environment of the periplasmic space, proteins that contain one or more disulfide bonds are best secreted. The presence of an N-terminal signal sequence appears to be necessary but not sufficient to direct a target protein to the periplasm. Translocation across the outer membrane and into the growth medium is inefficient. In most cases target proteins found in the growth medium are the result of damage to the cell envelope and do not represent true secretion (Stader and Silhavy, 1990). Translocation across the inner cell membrane of E. coli is incompletely understood (reviewed by Wickner, Driessen, and Hartl, 1991), and the efficiency of export will depend on the individual target protein. Currently the export cannot be predicted based on protein sequence, although some generalizations have been made about the sequence immediately following the signal peptide (Boyd and Beckwith, 1990; Yamane and Mizushima, 1988). Therefore it is possible to find target proteins in the cytoplasm (with uncleaved signal sequence) or in the periplasm in partially processed form, in place of or in addition to the expected periplasmic processed species. In some cases the proportion of protein that is exported can be increased by lowering the temperature 15 to 30°C during induction.
Post-translational Modification
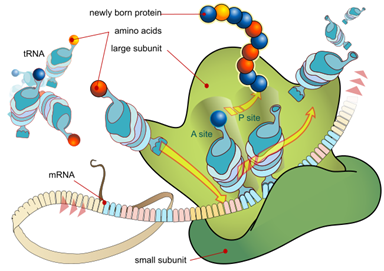 E. coli does not glycosylate or phosphorylate proteins or recognize proteolytic processing signals from eukaryotes, so take this into account when designing the cloning strategy. If proteolytic processing is needed, it is best to express only the coding sequences for the fully processed protein. If the protein of interest requires glycosylation for activity, and full activity is important in the final use, consider a eukaryotic host, such as Pichia, insect cells, or mammalian cells.
E. coli does not glycosylate or phosphorylate proteins or recognize proteolytic processing signals from eukaryotes, so take this into account when designing the cloning strategy. If proteolytic processing is needed, it is best to express only the coding sequences for the fully processed protein. If the protein of interest requires glycosylation for activity, and full activity is important in the final use, consider a eukaryotic host, such as Pichia, insect cells, or mammalian cells.
Is the Protein Potentially Toxic?
proteins might all be expected to have some toxic effect on E. coli. Expression of toxic proteins may be very low, and there will be strong selective pressure on cells to eliminate the gene of interest by point mutation to change the translation frame, insertion of a stop codon, or change in an amino acid residue critical to the protein’s function. Larger deletion of parts of the plasmid may also be seen. If there is a suggestion that the gene product will be toxic, use an gene expression vector with a tightly regulated promoter (e.g., T7, pETvectors). Minimize propagation of the cells to avoid opportunities for mutation and recombination.
Must Your Protein Be Functional?
 Each requirement placed on a recombinant protein will affect the choice of expression system. If a protein is to be used only to prepare antibody, it need not be soluble or active, and the production of inclusion bodies (aggregates of improperly folded protein) in E. coli may be all that is needed. Alternatively, if a protein’s biological activity will be assayed, or if it is to be used in structural studies (NMR, crystallography, etc.), a properly folded and soluble form will be required.
Each requirement placed on a recombinant protein will affect the choice of expression system. If a protein is to be used only to prepare antibody, it need not be soluble or active, and the production of inclusion bodies (aggregates of improperly folded protein) in E. coli may be all that is needed. Alternatively, if a protein’s biological activity will be assayed, or if it is to be used in structural studies (NMR, crystallography, etc.), a properly folded and soluble form will be required.
Will Structural Changes (Additional or Fewer Amino Acids) Affect Your Application?
structural studies or precise comparisons to a native protein are to be done, it is wise to eliminate amino acids added by cloning steps. PCR amplification is the most commonly used method to generate inserts for gene expression, and proper design of PCR primers can eliminate most or all additional residues in the protein.
RECOMBINANT PROTEIN EXPRESSION VECTORS
What Levels of Protein Expression Should You Expect?
 There are several systems available for protein expression in mammalian, insect, yeast, and E. coli. While it is impossible to predict the yields of protein from these systems for any given protein, some rough guidelines can be given. For any vector it is possible that no expression will be seen! Reported yields in stably transfected mammalian cells are in the range of 1 to 100mg/106 cells. Insect cell systems will yield between 5 and 200mg/L of culture (Schmidt et al., 1998), Pichia can produce up to 250mg/L (Eldin et al., 1997), and reported yields in E. coli range from 50mg to over 100mg/L. Usually yields of from 1 to 10mg/L can be expected from E. coli. Higher yields, up to a gram or more per liter, can be had using fermentation vessels where oxygen and pH levels can be controlled throughout the cell growth. The abovementioned values are guidelines; they are entirely dependent on the protein to be expressed. It is always best to test one or more systems in parallel to select the best solution. Nonbiological synthesis of protein is now possible as an alternative to production in a host organism (Kochendoerfer and Kent, 1999). Oligopeptides are synthesized and then assembled by chemical ligation to give full-length protein. The method has the potential to synthesize gram quantities of >30kDa proteins, and such preparations would of course be free of host contaminants that might interfere with function or use in diagnostic or therapeutic applications. Unfortunately, chemical synthesis of proteins is not widely available.
There are several systems available for protein expression in mammalian, insect, yeast, and E. coli. While it is impossible to predict the yields of protein from these systems for any given protein, some rough guidelines can be given. For any vector it is possible that no expression will be seen! Reported yields in stably transfected mammalian cells are in the range of 1 to 100mg/106 cells. Insect cell systems will yield between 5 and 200mg/L of culture (Schmidt et al., 1998), Pichia can produce up to 250mg/L (Eldin et al., 1997), and reported yields in E. coli range from 50mg to over 100mg/L. Usually yields of from 1 to 10mg/L can be expected from E. coli. Higher yields, up to a gram or more per liter, can be had using fermentation vessels where oxygen and pH levels can be controlled throughout the cell growth. The abovementioned values are guidelines; they are entirely dependent on the protein to be expressed. It is always best to test one or more systems in parallel to select the best solution. Nonbiological synthesis of protein is now possible as an alternative to production in a host organism (Kochendoerfer and Kent, 1999). Oligopeptides are synthesized and then assembled by chemical ligation to give full-length protein. The method has the potential to synthesize gram quantities of >30kDa proteins, and such preparations would of course be free of host contaminants that might interfere with function or use in diagnostic or therapeutic applications. Unfortunately, chemical synthesis of proteins is not widely available.
Which E. coli Strain Will Provide Maximal Protein Expression for Your Clone?
The choice of an expression host depends on the promoter system to be used. Promoters that depend on E. coli RNA polymerase can be expressed in most common cloning strains, while T7 promoter vectors must be used in E. coli that co-express T7 RNA polymerase (e.g., strains that contain the DE3 lysogen) (Dubendorff and Studier, 1991). Strains that are protease deficient (Bishai, Rappuoli, and Murphy, 1987) or overexpress chaparones have been shown to be useful for some proteins (Georgiou and Valax, 1996; Gilbert, 1994). At a minimum, a recombination deficient strain is advisable.Vendors of the commercially available E. coli expression vectors generally will recommend a host for use in expression. As with many questions related to protein expression, the results will depend on the nature of the protein of interest.A given gene may give high yields of intact protein in most strains, while the next would show no product except in a protease deficient host.
Why Should You Select a Fusion System?
Increased Yields
There are several reasons that one would choose to use a fusion system. Translational initiation from the amino terminal fusion partner may be more efficient than the start contributed by the protein of interest, so larger amounts of protein can be obtained as a fusion. In addition smaller proteins (<20kDa), or subfragments of larger ones often benefit from association with a stable fusion partner, due in part to improved folding or protection from proteolysis. Fusion with GST, MBP, and thioredoxin may be useful for this purpose.
Simplified Purification and Detection
Most of the commonly available fusion partners double as affinity tags, and these make isolation of the protein of interest relatively simple. Protein can often be purified to >90% in a single step. In contrast to conventional chromatographic techniques, little or no information about the sequence, pI, or other physical characteristics of the protein is needed in order to perform the purification. Novice chromatographers or those who have not developed methods for purification of the native protein are advised to begin with an affinity system. Detection of fusion proteins is a simple matter, since antibodies and colorimetric substrates are available for several of the more common fusion partners. Thus, if there is no established method to detect the protein, detection of the fusion partner can be the most convenient way to assay for the presence of the protein in cells and throughout purification and assay of the protein of interest.
When Should You Avoid a Fusion System?
Since affinity tags make purification relatively simple, and tags can be removed by proteolyic cleavage, use of a tag usually makes sense. If, on the other hand, a nonfusion vector has been used in earlier work, and one wishes to compare results with older data, use the nonfusion system. If there is an established method for purification and a biochemical assay or antibody available to detect the protein of interest, an affinity partner or tag for detection may simply be unnecessary. Ask again what use the protein will be put to. If the end application is likely to be sensitive to the presence of the tag (e.g., NMR, crystallography, therapeutics), and other conditions above are met, there is reason to avoid the tag. If a fusion affinity tag is desired, several are available.
 Susceptibility To Cleavage Enzymes
Susceptibility To Cleavage Enzymes
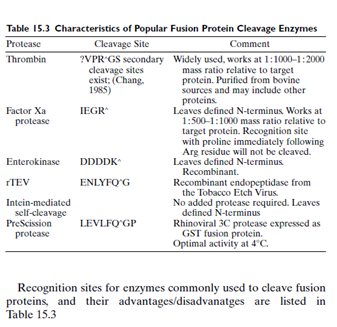
As discussed below, some fusion systems allow for the removal of the affinity tag by specific proteolytic or chemical cleavage. Before beginning any experiment, examine the sequence of the protein to be cloned and expressed. The protein of interest may have a binding site for one of the proteases listed in Table 15.3, and if so, this site should be avoided, or a different protein expression system might be required. Most proteases used for cleavage of fusion protein are quite specific, with theoretical frequencies of 10-6. However, it is best to check as a matter of course.
For many proteins, cleavage is not needed. If the goal of the work is to raise an antibody, the whole fusion protein can be used successfully as antigen-provided that antibodies to the tag do not interfere in the application. If, on the other hand, the protein is to be used in structural studies, or where the function of recombinant protein will be compared
Is It Necessary to Cleave the Tag off the Fusion Protein?
with native protein, it may be necessary to remove the fusion tag. Systems have been developed that use chemical (Nilsson et al., 1985) or specific proteolytic cleavage to separate the protein of interest from the fusion tag. The proteases have the advantage that cleavage is done at near neutral pH and at 4 to 37°C. In addition to proteolytic cleavage, the use of self-splicing inteins has been developed and commercialized by New England Biolabs. In this latter case fusion proteins with chitin-binding domain are bound to high molecular weight chitin chromatography media and incubated in the presence of a reducing agent, generally overnight. Protein splicing takes place, leaving the protein of interest in the flow through, while chitin and the spliced peptide remain bound.
Will Extra Amino Acid Residues Affect Your Protein of Interest after Digestion?
Depending on the protease, and the way in which the protein of interest was cloned in the expression vector, there may be one or more nonnative residues left at the amino terminal of the protein of interest following cleavage.Whether or not this poses a problem depends entirely on the protein and the use to which it will be put. Even the most demanding applications may not be negatively affected by the presence of extra amino terminal residues.Wherever possible, it is best to design a cloning strategy that at least minimizes the number of these residues, and if relatively inoccuous residues (e.g., glycine, serine) can be introduced, all the better.
©2013 BiologicsCorp, All right reserved.
Contact Us


