Protein Expression and Purification

Overview of protein expression and purification
What do you know about your protein
How to improve the expression level of active and soluble protein
Strategies for native protein and recombinant protein purification
Methods for protein seperation and protein purification
Custom Protein Service & Contact Us

1. Introduction
Bacterial expression is the most common expression system employed for the production of recombinant proteins. The organism, especially Escherichia coli (E. coli), is easy to manipulate, inexpensive in culturing and fast in generation of a recombinant protein. However, since it is a prokaryotic based system, heterologous eukaryotic proteins expressed are not correctly modified, and it can also be difficult to facilitate the secretion of expressed protein in large amounts. Moreover, proteins expressed in large amounts might precipitate, forming inclusion bodies and large complex proteins could be difficult to propagate.
2. Strategies for improving the expression level of active and soluble protein
2.1 FACTORS AFFECTING BACTERIAL EXPRESSION SYSTEM
2.1.1 Expression vectors design consideration
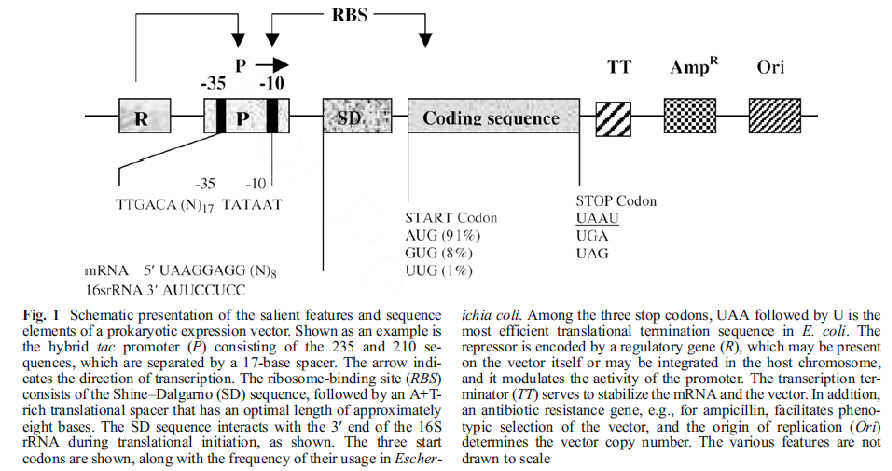
The construction of an expression plasmid requires several elements whose configuration must be carefully considered to ensure the highest levels of protein synthesis. The essential architecture of an E. coli expression vector is shown in (Fig. 1). The promoter is positioned approximately 10 bp to 100 bp upstream of the ribosome-binding site (RBS) and is under the control of a regulatory gene, which may be present on the vector itself or integrated in the host chromosome. Promoters of E. coli consist of a hexanucleotide sequence located approximately 35 bp upstream of the transcription initiation base (235 region),separated by a short spacer from another hexanucleotide sequence (210 region). There are many promoters available for gene expression in E. coli, including those derived from gram-positive bacteria and bacteriophages. A useful promoter exhibits several desirable features: it is strong, it has a low basal expression level (i.e., it is tightly regulated), it is easily transferable to other E. coli strains to facilitate testing of a large number of strains for protein yields, and its induction is simple and cost-effective. Downstream of the promoter is the RBS, which spans a region of approximately 54 nucleotides bound by positions 235 and 119-122 of the mRNA coding sequence. The Shine每Dalgarno (SD) site interacts with the 39 end of 16S rRNA during translation initiation. The distance between the SD site and the start codon ranges from 5 to 13 bases, and the sequence of this region should eliminate the potential of secondary-structure formation in the mRNA transcript, which can reduce the efficiency of translation initiation. Both 5∩and 3∩ regions of the RBS exhibit a bias toward a high-adenine content. The transcription terminator is located downstream of the coding sequence and serves both as a signal to terminate transcription and as a protective element composed of stemloop structures, protecting the mRNA from exonucleolytic degradation and extending the mRNA half-life. In addition to the above elements that have a direct impact on the efficiency of gene expression, vectors contain a gene that confers antibiotic resistance on the host to aid in plasmid selection and propagation. Finally, the copy number of plasmids is determined by the origin of replication. In specific cases, the use of runaway replicons results in massive amplification of plasmid copy number concomitant with higher yields of plasmid-encoded protein. The final rate of protein synthesis will normally depend on several factors: gene dosage, promoter strength, mRNA stability and the efficiency of translation initiation.
2.1.2 Codon usage/bias
Most of amino acids have been encoded by more than one codon and all available amino acid codons are bias utilized by each organism. Transfer RNA (tRNA) of cells reflects the codon bias of its mRNA. Observation of codon usage in E. coli reveals that highly expressed genes exhibit greater codon bias than poorly expressed ones and the frequency of synonymous codons used reflects the abundance of their cognate tRNAs. These can imply that heterologous genes with abundant codons, rarely used in E. coli, may not be efficiently expressed in E. coli and may lead to translation error. Codon bias becomes highly prevalent problems when rare codons in the transcripts form clusters such as doublets or triplets accumulation that is large in quantities.Translation error arised from rare codon bias includes mistranslational amino acid substitutions, frameshifting events or premature translational termination.
2.1.3 Protein folding
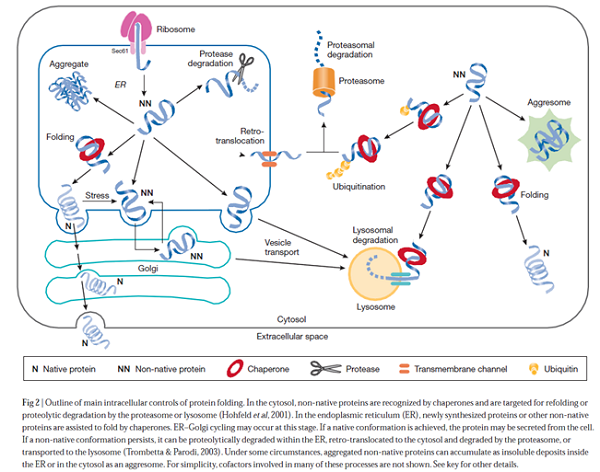 Expression of recombinant proteins in E. coli is mainly directed to three different locations i.e. the cytoplasm, the periplasm, and the growth medium (through secretion). Expression in the cytoplasm is preferred since the production yields are usually high. Cytoplasmic folding is often enhanced at low temperatures thus the use of cold-this is often accompanied by misfolding and segregation into insoluble aggregates known as inclusion bodies. Aggregation can be reduced to minimum through the control of parameters such as temperature, expression rate and host metabolism. Though formation of inclusion body renders easier protein purification, there is no guarantee that the in vitro refolding will generate large amounts of biologically active products. To release recombinant proteins into the periplasm and the growth medium, many systems have been studied. As such a approach is complicated, the systems have not been commercialized.
Expression of recombinant proteins in E. coli is mainly directed to three different locations i.e. the cytoplasm, the periplasm, and the growth medium (through secretion). Expression in the cytoplasm is preferred since the production yields are usually high. Cytoplasmic folding is often enhanced at low temperatures thus the use of cold-this is often accompanied by misfolding and segregation into insoluble aggregates known as inclusion bodies. Aggregation can be reduced to minimum through the control of parameters such as temperature, expression rate and host metabolism. Though formation of inclusion body renders easier protein purification, there is no guarantee that the in vitro refolding will generate large amounts of biologically active products. To release recombinant proteins into the periplasm and the growth medium, many systems have been studied. As such a approach is complicated, the systems have not been commercialized.
2.1.4 Protein phosphorylation and glycosylation
E. coli has limited eukaryotic post-translational machinery function, which is considered as a key disadvantage for producing the eukaryotic phosphoproteins i.e. serine/threonine/tyrosine protein kinases. To overcome these obstacles, co-expression of modified mammalian enzymes such as protein methylases and acetylases and their substrates from single or two separate plasmid vectors in the same E. coli may result in the production of recombinant proteins that closely resemble native eukaryotic proteins. Glycosylation is another complex process of posttranslational modification. It is responsible for the formation of cellular glycans which are often attached to proteins and lipids. Glycosyltransferase and glycosidases are enzymes responsible for glycosylation of many proteins. Glycoproteins, which are commonly distributed in eukaryotic cells, are rarely presented in prokaryotic organisms because cellular organelles essential for glycosylation are missing in these organisms.
2.1.5 Stability of mRNA
The stability of mRNA affects gene expression rates. The average half-life of mRNA in E. coli at 37 ranges from seconds to the maximum at 20 min and the expression rate depends directly on the inherent stability of mRNA.Degradation of mRNA by RNases can be protected through RNA folding, ribosomes and stability modulation by polyadenylation. Recombinant protein expression systems with mRNA stability enhancement is commercially available, for example, BL21 star strain, containing a mutation in the gene encoding RNaseE.
2.1.6 Promoter strength
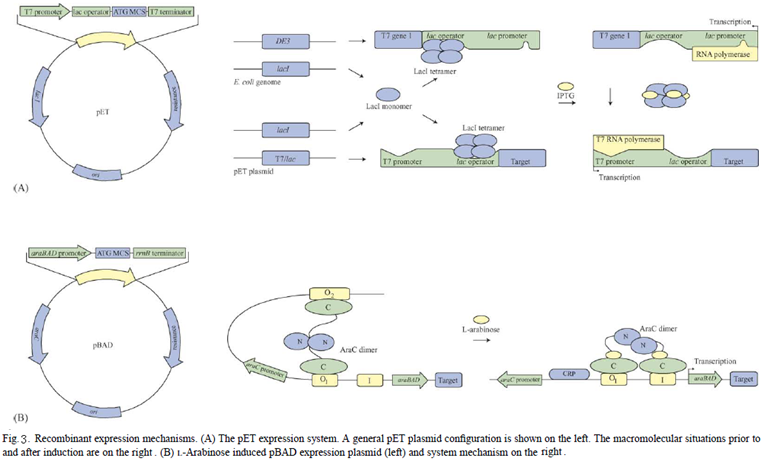
Recombinant expression plasmids require strong transcriptional promoter to enable high-level gene expression. Promoter must be induced using either thermal or chemical means and the most common inducer is the sugar molecular isopyl-beta-D-thiogalactopyranoside (IPTG). However, IPTG is not suitable for large scale production of human therapeutic proteins because it is toxic and expensive.
2.2 TIGHT CONTROL OF THE E. COLI CELLULAR MILIEU
Expression of soluble proteins can be regulated through many factors that the host cell normally use in controlling of toxic protein expression.
2.3 MODIFICATION OF E. COLI HOST STRAIN
The strain or genetic background of host strain is important for recombinant protein expression. Expression strains should be deficient in harmful proteases, but should stably maintain the expression plasmid and confer the relevant genetic elements to the expression system. E. coli BL21( DE3) is an example of the most common host and it has been proven outstanding in application for standard recombinant expression. It can grow efficiently in minimal media as nonpathogenic bacterium that cannot survive to cause diseases in host tissues.
2.4 MODIFICATION OF MEDIA COMPOSITION
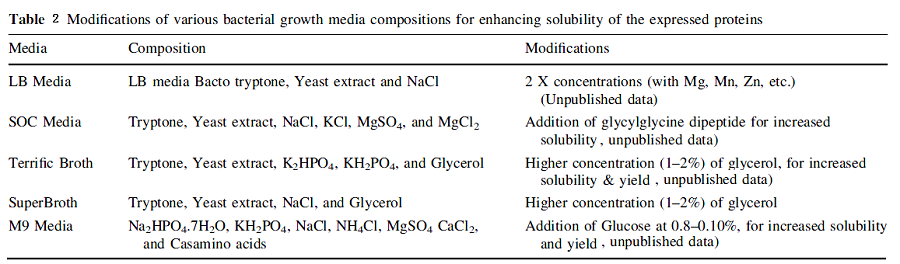
Production of recombinant protein requires nutrients for bacterial growth and there is a limited control on the growth parameters. This process often leads to changes in substrate depletion, pH, and concentration of dissolved oxygen as well as accumulation of inhibitory substances from various metabolic pathways. These changes are not beneficial for the production of either soluble or correctly folded active protein. Proper and efficient protein folding might require specific cofactors in the growth media such as metal ions.Addition of these essential factors to the culture media could considerably increase the yield as well as the folding rate of the soluble proteins.
2.5 EXPRESSION AT LOWER TEMPERATURES
Protein expression in E. coli growing at low temperature has shown its success in improving the solubility of proteins that are difficult to express as soluble proteins. Expression at low temperature conditions leads to the increase of stability and correct folding patterns due to the fact that hydrophobic interactions determining inclusion body formation are temperature dependent. Moreover, any expression associated with toxic phenotype observed at 37°C incubation conditions, will be suppressed at low temperatures. The increase of expression and activity of lower temperatures growth is associated with increased expression of chaperones in E. coli. Therefore, growth at a temperature range of (15-23)°C, could also lead to a significant reduction of expressed protein degradation.
2.6 CO-EXPRESSION OF MOLECULAR CHAPERONES
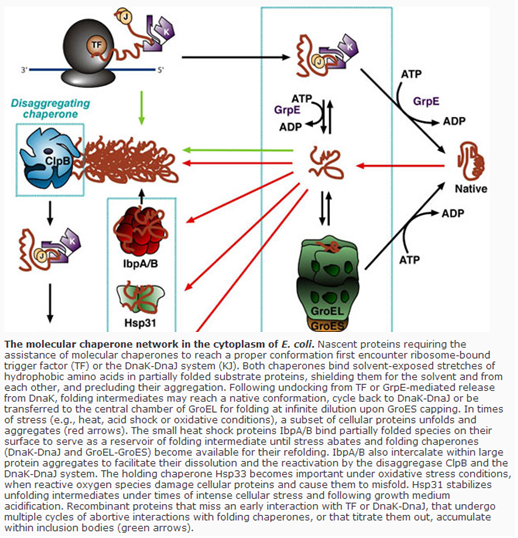
Molecular chaperones are proteins adapted to assist de novo protein folding and /or facilitate expressed polypeptide's proper conformation attainment. Coexpression of molecular chaperone strategy has been adopted for prevention of inclusion body formation, leading to improving of solubility of the recombinant protein. Chaperones are working as a trigger factor assisting in recombinant protein refolding. These polypeptides continue to attain folding into the native state even after their release from the protein-chaperone complex. Moreover, some chaperons could also prevent protein aggregation.
2.7 INCLUSION BODY FOLDING
Inclusion bodies are intracellular protein aggregates which were observed when the target gene is over expressed in the cytoplasm of E. coli. Formation of inclusion bodies in recombinant expression systems occurs as a result of erroneous equilibrium between in vitro protein solubilization and aggregation and might lead to unfavorable protein folding.
2.8 REFOLDING/RESOLUBILIZATION OF E. COLI INCLUSION BODY PROTEINS
Recombinant proteins expressed as inclusion bodies in E. coli have been widely used for the commercial product of therapeutic proteins. The major drawbacks during the refolding of inclusion body proteins into more efficient, soluble and correct folded product are reducing of recovery. Other than that the requirement for optimization of refolding conditions for each target protein and the resolubilization procedure could possibly affect the activity of refolded protein. Therefore, the production of soluble recombinant protein remains a preferable alternative than the in vitro refolding procedures.
2.9 ISOLATION/SOLUBILIZATION OF INCLUSION BODIES
Isolation of inclusion bodies can be done by lysozyme treatment along with EDTA before cell homogenization to facilitate cell disruption. Inclusion bodies are recovered by low speed centrifugation of bacterial cells that has been mechanically disrupted either by ultrasonication or high pressure homogenization. Bacterial cell envelop or outer membrane proteins may co-precipitate with the insoluble fractions as the inclusion body impurities. These contaminants can easily be removed by adding detergents such as Triton X-100 or low concentrations of chaotropic compounds. After removal of the impurities, inclusion bodies are solubilized by various concentrations of chaotropic agents such as urea or guanidinium hydrochloride. The latter is more favored due to its better chaotropic properties. Inclusion body proteins that were solubilized under mild denatured conditions are better in refolding yields and retaining of biological activities.
2.10 REFOLDING OF SOLUBILIZED AND UNFOLDED PROTEINS
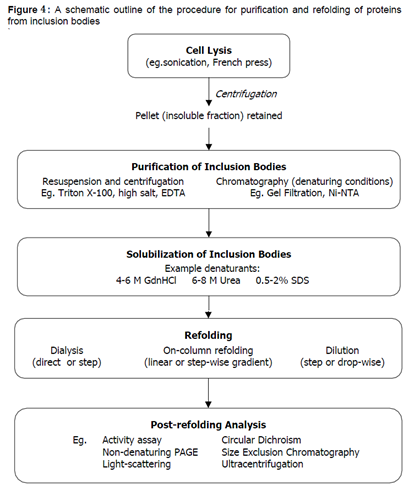
The methods normally used for solubilization of inclusion body could lead to non-native conformation of the expressed protein. This problem could be resolved by proper refolding procedures of target protein at low denaturant concentrations. Higher concentration of the unfolded protein often leads to decreased refolding yields, regardless of refolding method. So, it is desirable to keep the concentration of the initial un-folded protein to a minimum level if higher and correct refolding proteins are expected.
©2013 BiologicsCorp, All right reserved.
Contact Us


