Codon Optimization
Codon optimization is a novel technique to improve protein expression level in living organism by increasing translational efficiency of target gene. BiologicsCorp provides state-of-the-art algorithms to optimize gene sequences using in-house pre-computed software from a predicted group of highly expressed genes from thousands of samples. Generally, a codon adaptation index (CAI, a widespread technique for codon usage bias analysis) of less than 0.8 calculated by our online CAI calculator is considered as codon optimization needed.
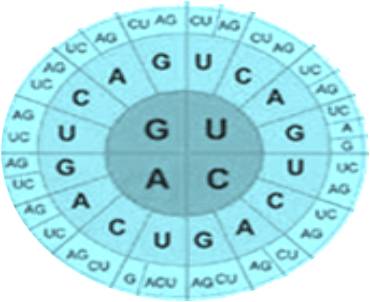
Codon optimization parameters

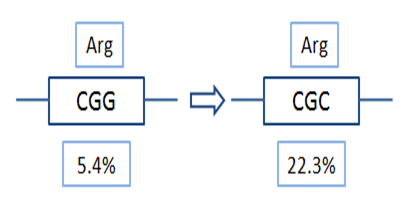
Codon usage bias
The technical platform changes rare codons to match the most prevalent tRNAs depend on specific species. It prevents protein synthesis rate slowdown and low expression yield cause by deficient particular aminoacyl-tRNA and corresponding codons. For certain DNA sequences, our rare codon analyzer is available to evaluate your projects.
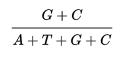
GC content
GC pair is bound by three hydrogen bonds, which improve DNA stability. For certain gene sequences, the GC-content can be accurately calculated by our online GC content calculator.
mRNA structure
mRNA structure has critical roles in translation regulation process. Our codon optimization software opens intra-molecularly formed stems and subsequently increases protein expression yield by extending mRNA stability.
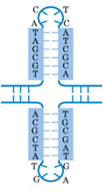
Repeated sequences
Repeated sequences are patterns of nucleic acids occur in multiple copies throughout the genome. Its redundancy causes difficulty in gene synthesis and protein expression. Reverse complement repeats have significantly impact on mRNA secondary structure as well.
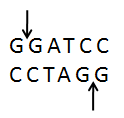
Avoided restriction enzymes
Codons of restriction enzymes in the target sequence should be replaced by synonymous nucleotide in the process of codon optimization to eliminate restriction enzyme cleavage sites and facilitate DNA cloning operations.
Non-coding region
Non-coding DNA elements are also considered as important parts of codon optimization projects. Choosing optimal promoter and terminator combination may lead to high expression level of your proteins of interest.
Motifs
- TATA box: a DNA sequence found in the promoter region of genes and is involved in the process of transcription.
- SD sequence: a ribosomal binding site in prokaryotic mRNA to trigger protein synthesis process.
- Kozak sequence: a sequence on eukaryotic mRNA and contribute to initiation of the translation process.
- Chi site: work as stimulators of DNA double strand break repair in bacteria.
- Cryptic splice sites: utilized in RNA splicing and induce cryptic splicing.
Codon optimization sample data
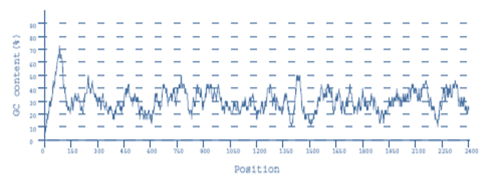
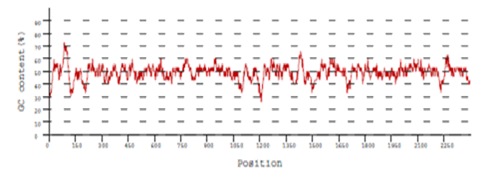
Recent publications citing BiologicsCorp’s codon optimization tools:
- Ángela Patricia Guerra, et al. "Production of recombinant proteins from Plasmodium falciparum in Escherichia coli." Biomédica Revista Del Instituto Nacional De Salud 36(2016).
- Agarwal, Shivangi, et al. "Induced autoprocessing of the cytopathic Makes Caterpillars Floppy-like effector domain of the Vibrio vulnificus, MARTX toxin." Cellular Microbiology 17.10(2015):1494–1509.
Codon optimization case study
Our codon optimization tools modified synonymous codons and mRNA stem-loop structure to subsequently increase protein expression level.
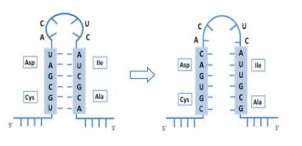
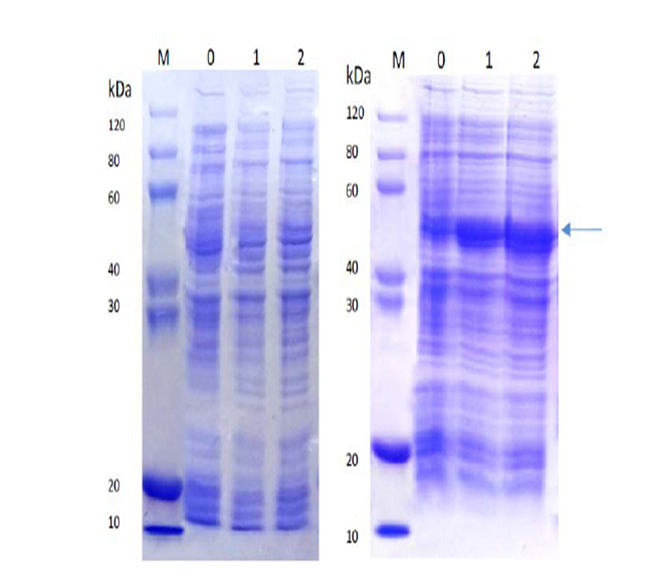
Lane M: Protein marker
Lane 0: Cell lysate without induction
Lane 1: Cell lysate with IPTG induction at 15°C, overnight
Lane 2: Cell lysate with IPTG induction at 37°C, 4h
Contact Us


